Section: New Software and Platforms
Shanoir
Keywords: Shanoir - Webservices - Biology - Health - DICOM - Neuroimaging - Medical imaging - PACS - Nifti
Functional Description SHAring NeurOImaging Resources (Shanoir, Previously InriaNeuroTk) is an open source software platform designed to structure, manage, archive, visualize and share neuroimaging data with an emphasis on multi-centric collaborative research projects. It provides common features of neuroimaging data management systems along with research-oriented data organization and enhanced accessibility (see figure 4 ).
Shanoir is a secured J2EE application running on a JBoss server, reachable via graphical interfaces in a browser or by third party programs via web services. It behaves as a repository of neuroimaging files coupled with a relational database hoding meta-data. The data model, based on OntoNeurolog, an ontology devoted to the neuroimaging field, is structured around the concept of "research study". A research study includes patients who themselves have examinations that either produce image acquisitions or clinical scores. Each image acquisition is composed of datasets represented by their acquisition parameters and image files. The system only keeps anonymous data.
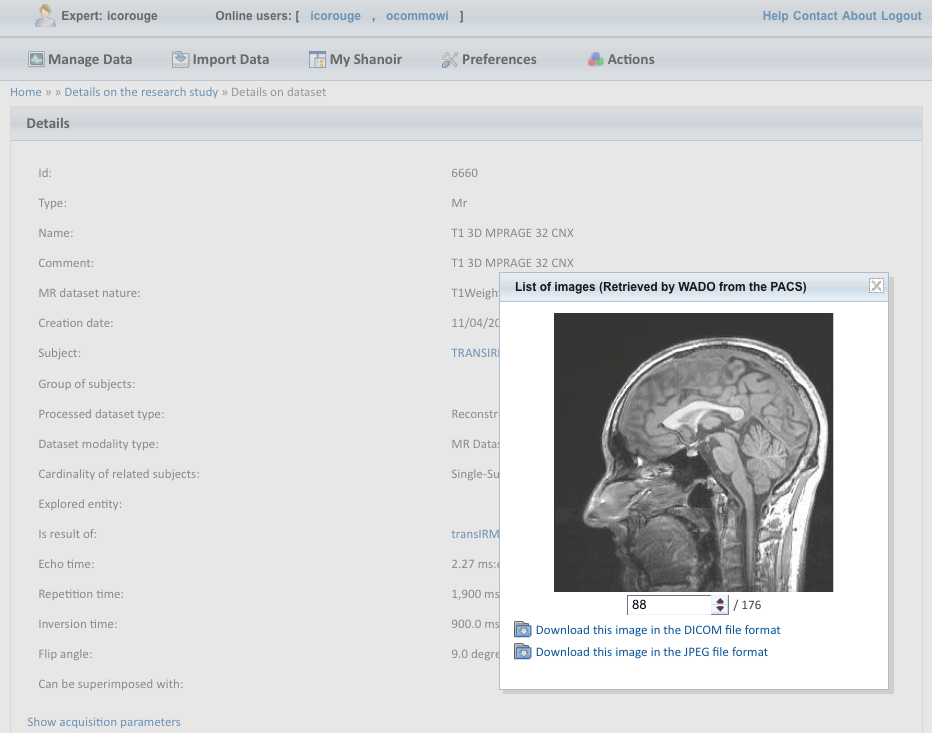
Image file imports are possible from various sources (DICOM CDs, PACs, image files in NIfTI / Analyze format) using either online wizards, with completions of related meta-data, or commande line tools. Once de-identified during the import phase, DICOM header's customizable feature. Shanoir can also record any executed processing allowing to retrieve workflows applied to a particular dataset along with the intermediate data.
The clinical scores resulting from instrument based assessments (e.g. neuropsychological tests) can also be entered and easily retrieved and exported in different formats (Excel, CSV, Xml). Scores and image acquisitions are bound together which makes relationship analysis possible. The instrument database is scalable and new measures can be added in order to meet specific project needs, by use of intuitive graphical interfaces.
Using cross-data navigation and advanced search criteria, the users can quickly point to a subset of data to be downloaded. Client side applications have as well been developed to illustrate how to locally access and exploit data through the available web services. With regards to security, the system requires authentication and user rights are tunable for each hosted studies. A study responsible can thereby define the users allowed to see, download or import data into his study or simply make it public.
Shanoir serves neuroimaging researchers in organizing efficiently their studies while cooperating with other laboratories. By managing patient privacy, Shanoir allows the exploitation of clinical data in a research context. It is finally a handy solution to publish and share data with a broader community.
Shanoir integrates the enterprise search platform, Apache Solr, to provide the users a vast array of advanced features such as near real-time indexing and queries, full-text search, faceted navigation, autosuggestion and autocomplete.